Modern Birds: The Neornithes
|
The Neornithes are defined as the smallest clade (an ancestral population and all of its descendents) that contains all currently living birds.
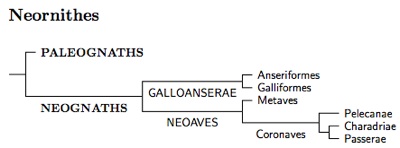
One problem that crops up when using genetic methods to study the early history of Neornithes, is that random DNA variation creates a lot of noise, and the signal-to-noise ratio is often unfavorable. This has caused problems, but they have been solved (e.g., Braun and Kimball, 2002) and the early history of Neornithes is now uncontroversial. The most basal (oldest) division in the tree of living birds has been identified. It is the division between the Paleognaths (Ostrich, Rheas, Cassowaries, Emus, Kiwis, Tinamous) and the rest of the living birds (Neognaths). The names Paleognath (old jaw) and Neognath (new jaw) reflect differences in the skulls of the two types of bird.
The split may have occurred during the early Cretaceous, more than 100 million years ago. The Paleognaths also appear in the Sibley-Monroe list. More recent evidence has strengthened the notion that the Paleognaths form a natural group, and that they include the recently extinct Elephant Birds of Madagascar (Aepyornithidae) and Moas of New Zealand (Dinornithidae), and not so recently extinct birds such as the Lithornidae. However, the traditional division of the Paleognaths into the flightless Ratites and volant Tinamous has recently been called into question (Harshman et al., 2008).
Harshman et al. suggest that flightlessness has evolved in the Paleognaths at least three times, and probably four: ostriches, rheas, cassowaries/emus/kiwis/elephant birds, and probably separately in the moas. The taxonomy here is based on Harshman et al. (2008) and Hackett et al. (2008).
In fact, there is further relevant DNA evidence indicating that the extinct elephant birds of Madagascar are the closest relatives of the kiwis (Mitchell et al., 2014b), with a common ancestor perhaps 50 million years ago. This is a problem for vicariance theories because Madagascar (and India) likely separated from Gondwana considerably earlier, perhaps 120 million years ago. In that case, the ancestor of the elephant birds would have had to fly, and we can infer that flight was indpendently lost in the rheas, emu/cassorwary, elephant bird, and kiwi lineages, a total of 6 losses of flight. This is not an unreasonable number. One only has to consider all of the flightless rails to see that.
Galloanserae
Following the publication of Sibley and Ahlquist's book (1990), ornithologists have gradually realized that the deepest division in the Neognaths separates the waterfowl and gallinaceous birds (Galloanserae) from everything else (Neoaves). This is also now uncontroversial. These divisions are recognized in recent checklists such as the current versions of the AOU, ABA, Howard-Moore (3rd and 4th ed.), and Gill and Donkser's IOC World Bird List. In contrast, the older modified Sibley-Ahlquist-Monroe list of Gill (1995) places the Anseriformes after the Pelecaniformes with the Phoenicopteriformes, Ciconiiformes, and Falconiformes separating them from the Galliformes.
The Galloanserae in turn divide into the Anseriformes and Galliformes. One of the Anseriform families is the extinct Presbyornithidae. I've included them in the detailed tree diagram so you can see how these numerous and well-known fossils fit in a modern taxonomy (Ericson, 1997). The oldest Presbyornis fossils date back about 60 million years.
Neoaves
We conclude that avian mitochondrial genomes reject the hypothesis of a shared evolutionary history for hummingbirds, kagu, tropicbirds and flamingos.—Morgan-Richards et al., 2008
Once we get to Neoaves, things become more contentious. The diversification of Neoaves appears to have happened very quickly (e.g., Poe and Chubb, 2004). This means we have lots of noise relative to the signal. Not surprisingly, the higher taxonomy of Neoaves remains unsettled, with different researchers coming to different conclusions. However, most recent papers fall into two major groups, those based solely on mitochondrial DNA (e.g., Harrison et al. 2004; Slack et al. 2007; Gibb et al. 2007; Morgan-Richards et al. 2008; Pratt et al. 2009; Pacheco et al. 2011), and those that also rely on nuclear DNA including β-fibrinogen (Fain and Houde, 2004; Ericson et al. 2006; Hackett et al., 2008; Pásko et al., 2011). There are some other results around that don't fit comfortably with either version (e.g., Sorenson et al., 2003). The two main groups of studies yield incompatible topologies (see Morgan-Richards et al., 2008).
Which then is correct? The short answer is that we don't know. However, the mtDNA phylogenies are not currently suitable for my purposes. They are too incomplete, and keep changing as they add taxa. It's not just the more problematic taxa that are missing. They simply do not provide a consistent picture. The most comprehensive mitochondrial analysis is that of Pacheco et al. (2011). Yet they do not include the Gruiformes and they threw out the Cathartidae. A number of other orders are not included either. Recent mitochondrial studies have also shown some odd results at lower levels—paraphyletic shorebirds in Pratt et al.; the potoos grouped with ibises and pelicans in Pacheco et al. (see their Figure 1B, which has been arranged in a way that obscures this).
The combined nuclear/mitochondrial papers suffer from neither of these problems. Both Ericson et al. (2006) and Hackett et al. (2008) cover most taxa of interest and deliver generally consistent results. They are also consistent with the de novo reanalysis of Pasko et al. (2011), and the more restricted studies by Suh et al. (2011) and Wang et al. (2011). They are similar to the analysis of McCormack et al. based on ultraconserved elements (McCormack et al., 2013) and are also consistent with the early RAG-1 study by Groth and Barrowclough (1999).
However, the combined studies are somewhat suspect at the highest levels (can Metaves really be true??). Although it has been claimed that their results are purely due to the 7th intron of the beta-fibrinogen gene, that's not entirely true. The 5th intron gives similar but different results (I don't know if the differences are real or whether it means that neither is a true phylogenetic signal). Even without β-fibrinogen, some of the big differences are still there (see Figure ESM-6 in the supplementary material of Ericson et al., 2006).
There's also an interesting paper by Nabholtz et al. (2011) that suggests differences in base composition are causing problems for the mitochondrial studies. They don't look at the same genes and use only a small set of taxa, so it is impossible to say if this might lead to some reconciliation of the two major approaches.
The bottom line is that the only high-level molecular phylogenies that are both complete and reasonable at lower levels are the very similar Ericson and Hackett trees. That's why I use them. That doesn't mean I'm entirely convinced by them, only that I have more issues with the alternatives. If I picked a different arrangement, I'd probably base it on Ericson et al.'s ESM-6 or Cracraft et al. (2004).
Troublesome Taxa
We can group the families in Neoaves into a reasonable collection of orders (46 in this classification, 39 of them in Neoaves), but correct placement of over a third of the 39 orders in Neoaves remains uncertain. Eventually, we expect to be able to read enough DNA of enough species to untangle this, but for now, it is not possible to read and analyze the entire DNA of all of the birds. Until recently, the best DNA studies were either deep but narrow, including many genes from a few species, or are shallow and wide, including fewer genes from many more species. Hackett et al. (2008) changed that, using a dataset that set new standards in both depth and width. Even so, many of the major branches remain somewhat uncertainly positioned. They also provide the most complete analysis of the higher taxonomy of Neoaves, and generally reinforce and refine the results of Ericson et al. (2006a). Beginning with version 2.1, this taxonomy is used in these web pages. Key features of this arrangement depend on the analysis of a single gene, and may be subject to change.
Many of the 46 orders are traditional, but I have subdivided a few to ensure that they are monophyletic. I may eventually recombine some of the orders once it is clear that the phylogenetic tree allows it.
Five of the 46 orders are Paleognaths and two are Galloanseres, leaving 38 in Neoaves. Many of these can be placed in larger groupings with reasonable confidence, and these groups can be placed on the tree. The larger groups are Aequornithes (8 orders), Charadriae (1 order), and Passerae (13 orders). In the case of the Passerae, there is a core group that is supported by a wide variety of studies, and other orders that probably belong there too.
Those three groups account for 22 orders. The other 17 orders, containing about 1/9 of all living birds, are the “troublesome taxa”. They are troublesome because we are still uncertain, and sometime highly uncertain, about where they fit on the avian tree. We know they belong somewhere in Neoaves, but where remains a mystery. In TiF order, the troublesome taxa are:
- Mirandornithes
- Phoenicopteriformes (flamingos)
- Podicipediformes (grebes)
- Phaethontiformes (tropicbirds)
- Pterocliformes (sandgrouse)
- Mesitornithiformes (mesites)
- Columbiformes (pigeons and doves)
- Eurypygiformes (kagu and sunbittern)
- Strisores
- Steatornithiformes (oilbird)
- Nyctibiiformes (potoos)
- Podargiformes (frogmouths)
- Caprimulgiformes (nighthawks and nightjars)
- Apodiformes (owlet-nightjars, hummingbirds and swifts)
- Opisthocomiformes (hoatzin)
- Otidiiformes (bustards)
- Gruiformes (cranes and rails)
- Cuculiformes (cuckoos)
- Musophagiformes (turacos)
The 5 orders grouped as the Strisores are likely each other's closest relatives. Still, they are troublesome because there is considerable uncertainty about where the whole group goes. Moreover, genetic studies often fail to pull them together as a group, especially if β-fibrinogen is not included.
There's another pair we know something more about. The Phoenicopteriformes (flamingos) and Podicipediformes (grebes) are each other's closest relatives. Collectively, they are referred to as Mirandornithes. There's uncertainty about where Mirandornithes goes also. One possibility is that they are related to the shorebirds, but other evidence points elsewhere.
There is even more uncertainty attached to the remaining 10 troublesome taxa. We aren't even sure what their closest relatives are. One proposed solution to the problem of the troublesome taxa is the Metaves Hypothesis.